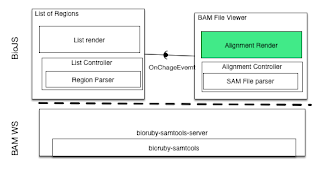
One of the key features when browsing BAM files is to be able to see the insertion, deletions, and SNPs. For that, not only displaying the base is important, but adding colours help to highlight the features. For that, I had to parse the CIGAR line to know the length of the read and the actual interoperation.
The orientation is stored in one of the flags (0x0010). To validate the orientation, I started adding a method to the sam line which tells if the orientation is forward or not. This makes the code later a bit clearer
has_flag : function (f){
f = parseInt(f);
return (this.flags & f) == f ;
},
forward: function(){
return this.has_flag(16);
}
Then, I got a function that paints the DIV with all the bases inside and applies a CSS style to each one of the divs for each base. The CSS can be edited externally and the functionality should stay. I have to say that I spent a lot of time testing this, as for some wired reason the CSS is not applied at the right time.
build_div: function(){
var new_div = document.createElement("div");
new_div.style.height = container.opt.fontSize ;
new_div.style.position = "absolute";
n_pos = ( this.pos - 1) * container.opt.base_width;
new_div.style.left = n_pos + "px";
if(this.forward()){
new_div.classList.add("bam_forward");
}else{
new_div.classList.add("bam_reverse");
}
var cigars = this.cigar.replace(/([SIXMND])/g, ":$1,");
var cigars_array = cigars.split(',');
var cig_index = 0;
this.len = 0
var cig_end = -1;
var cig ;
var key;
var length;
var cig_index = 0;
var last_div;
changed = true;
for ( var i = 0; i < this.seq.length; i++ ){
if(i > cig_end || changed == true){
cig = cigars_array[cig_index].split(":");
key = cig[1];
length = parseInt(cig[0]);
cig_end = i + length;
cig_index +=1
changed = false;
}
if(key == "M" || key == "X" || key == "="){
display_base = this.seq[i];
var current_base_span = document.createElement("div");
new_div.appendChild(current_base_span);
current_base_span.className = "bam_base_" + display_base;
current_base_span.style.width = container.opt.base_width + "px";
current_base_span.style.cssFloat = "left";
current_base_span.appendChild(current_base_span.ownerDocument.createTextNode(display_base));
last_div = current_base_span;
this.len += 1
current_base_span.id = this.len
}else if(key == "I"){
last_div.classList.add("bam_base_I");
changed = true;
}else if(key == "D" || key == "N"){
for (var j = 0; j < length; j ++ ) {
display_base = "*";
var current_base_span = document.createElement("div");
current_base_span.classList.add("bam_base");
current_base_span.classList.add("bam_base_D");
current_base_span.style.width = container.opt.base_width + "px";
current_base_span.style.cssFloat = "left";
current_base_span.appendChild(current_base_span.ownerDocument.createTextNode(display_base));
last_div = current_base_span;
new_div.appendChild(current_base_span);
this.len += 1;
current_base_span.id = this.len
}
changed = true;
//cig_index += 1;
i--;
}
}
new_div.style.width = container.opt.base_width * this.len + "px";
this.div = new_div;
}};